EnTAP required flags¶
--entap-annotation /path/to/your/final_annotation.tsv¶
Provide the path to the output of the protein annotation. The first column should be the name of a gene in the “g###.t1” format. As of this version, only braker 2.05 outputs annotated with ENTAP are recognized due to building resources. It will refuse to run with other formats.
The trailing t1 is disregarded and there should be no recognition issues if everything is run with the latest version of gFACs. Additionally, all ENTAP steps are done before fasta steps so all outputs will reflect filters that require the annotation.
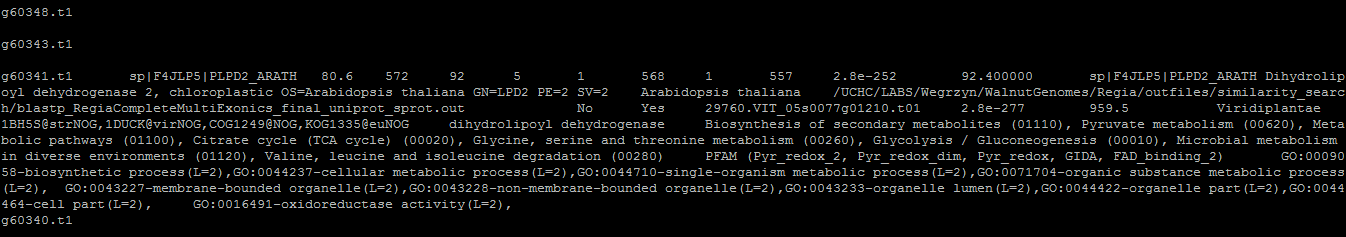
The annotation should look something like this:
--annotated-all-genes-only¶
Only genes that have an associated similarity search OR EggNOG annotation are kept. Done by the script task_scripts/annotated_all_genes_only.pl. Passing genes will remain in the gene table. Results of this filter are printed in the log.
--annotated-ss-genes-only¶
Only genes that have an associated similarity search annotation are kept. Done by the script task_scripts/annotated_ss_genes_only.pl. Passing genes will remain in the gene table. Results of this filter are printed in the log.