
Introduction to gFACs¶
gFACs is a filtering, analysis, and conversion tool to unify genome annotations across alignment and gene prediction frameworks. It was developed by Madison Caballero and Dr. Jill Wegrzyn of the Plant Computational Genomics Lab at the University of Connecticut.
Find gFACs on GitLab.
Version 1.1.2 - 07/17/2020
How to cite:
Comments? Questions? Email me at Madison.Caballero@uconn.edu
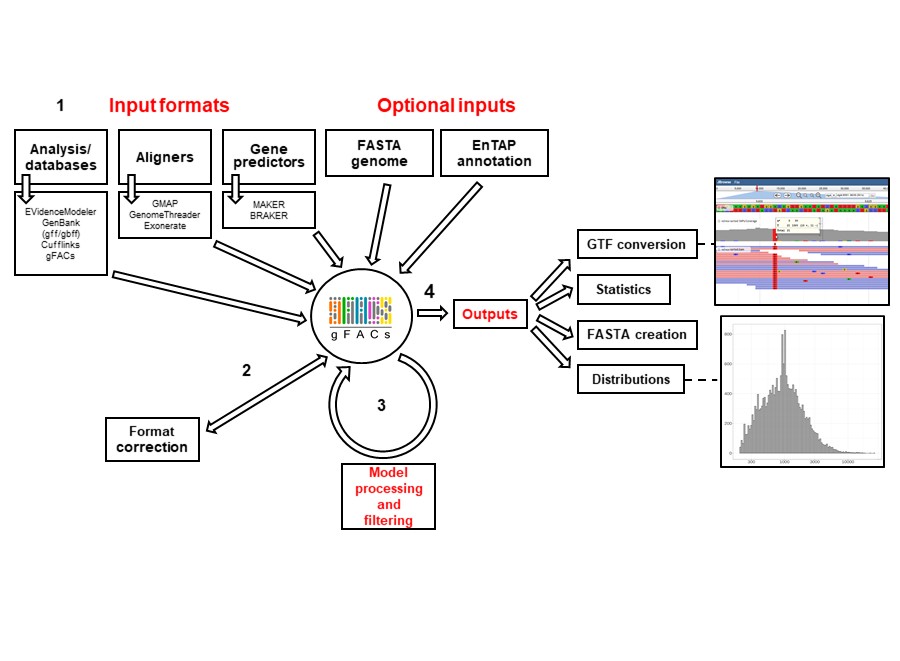
Getting started¶
Running gFACs¶
- Filter flags
- Basic flags
- -p [prefix]
- --false-run
- --no-processing
- --no-gene-redefining
- --rem-3prime-incompletes
- --rem-5prime-incompletes
- --rem-5prime-3prime-incompletes
- --rem-all-incompletes
- --rem-monoexonics
- --rem-multiexonics
- --min-exon-size [number]
- --min-intron-size [number]
- --min-CDS-size [number]
- --unique-genes-only
- --sort-by-chromosome
- EnTAP required flags
- Fasta required flags
- Basic flags
- Output flags
Support scripts¶