EnTAP required flags¶
--entap-annotation /path/to/your/final_annotation.tsv¶
Provide the path to the output of the protein annotation. The first column should be the name of a gene that matches the gene name in the gene table. It will run with other formats but I encourage using a gFACs format input (see FAQ for EnTAP run details). Use gFACs to filter an original annotation, functionally annotate with EnTAP, then use the gFACs output and EnTAP output to filter again.
All versions of EnTAP (including future versions) should be compatible.
If issues arise, contact me at the gFACs GitLab.
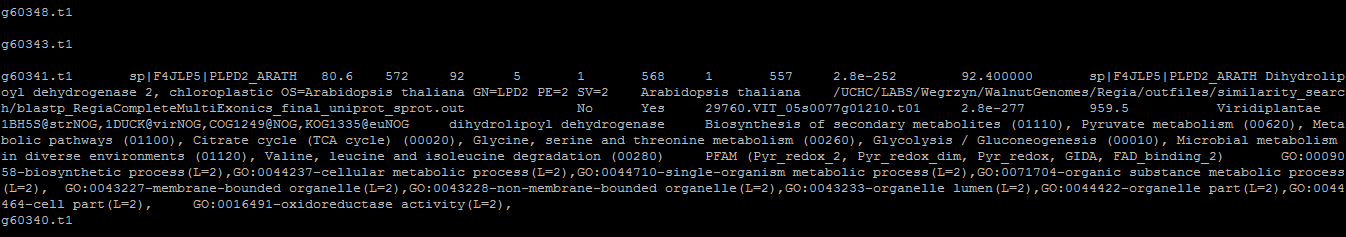
The annotation should look something like this:
--annotated-all-genes-only¶
Only genes that have an associated similarity search OR EggNOG annotation are kept. Done by the script task_scripts/annotated_all_genes_only.pl. Passing genes will remain in the gene table. Results of this filter are printed in the log.
--annotated-ss-genes-only¶
Only genes that have an associated similarity search annotation are kept. Done by the script task_scripts/annotated_ss_genes_only.pl. Passing genes will remain in the gene table. Results of this filter are printed in the log.